── Attaching core tidyverse packages ──────────────────────── tidyverse 2.0.0 ──
✔ dplyr 1.1.4 ✔ readr 2.1.5
✔ forcats 1.0.0 ✔ stringr 1.5.1
✔ ggplot2 3.5.1 ✔ tibble 3.2.1
✔ lubridate 1.9.4 ✔ tidyr 1.3.1
✔ purrr 1.0.2
── Conflicts ────────────────────────────────────────── tidyverse_conflicts() ──
✖ dplyr::filter() masks stats::filter()
✖ dplyr::lag() masks stats::lag()
ℹ Use the conflicted package (<http://conflicted.r-lib.org/>) to force all conflicts to become errors
── Attaching packages ────────────────────────────────────── tidymodels 1.2.0 ──
✔ broom 1.0.7 ✔ rsample 1.2.1
✔ dials 1.3.0 ✔ tune 1.2.1
✔ infer 1.0.7 ✔ workflows 1.1.4
✔ modeldata 1.4.0 ✔ workflowsets 1.1.0
✔ parsnip 1.2.1 ✔ yardstick 1.3.2
✔ recipes 1.1.0
── Conflicts ───────────────────────────────────────── tidymodels_conflicts() ──
✖ scales::discard() masks purrr::discard()
✖ dplyr::filter() masks stats::filter()
✖ recipes::fixed() masks stringr::fixed()
✖ dplyr::lag() masks stats::lag()
✖ yardstick::spec() masks readr::spec()
✖ recipes::step() masks stats::step()
• Dig deeper into tidy modeling with R at https://www.tmwr.org
An Example
library (modeldata) # Used for the data crickets names (crickets)
[1] "species" "temp" "rate"
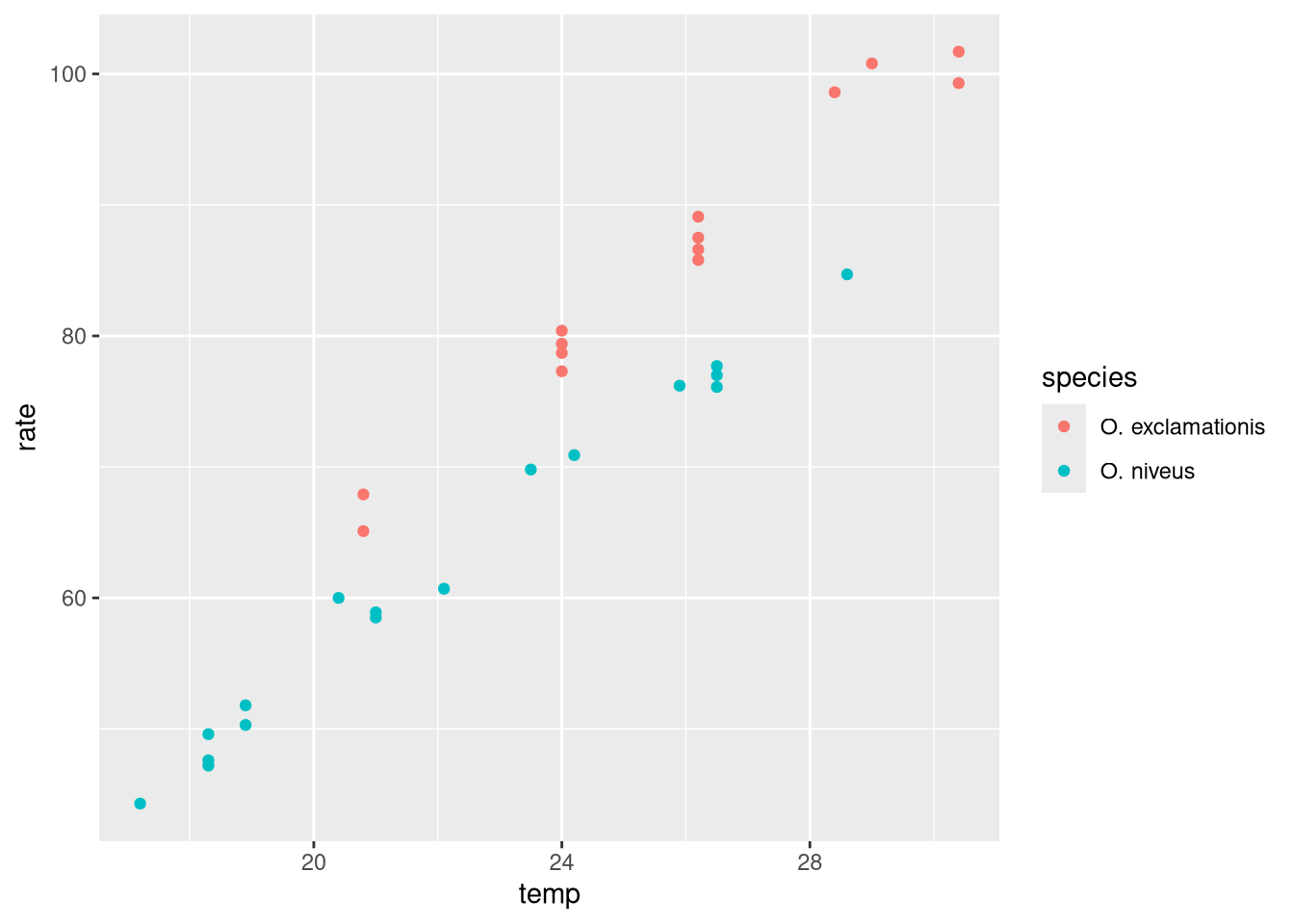
Plot the chirp rate on the y-axis and the temperature on the x-axis, colored by species.
ggplot (data = crickets, aes (x = temp, y = rate, color = species)) + geom_point ()
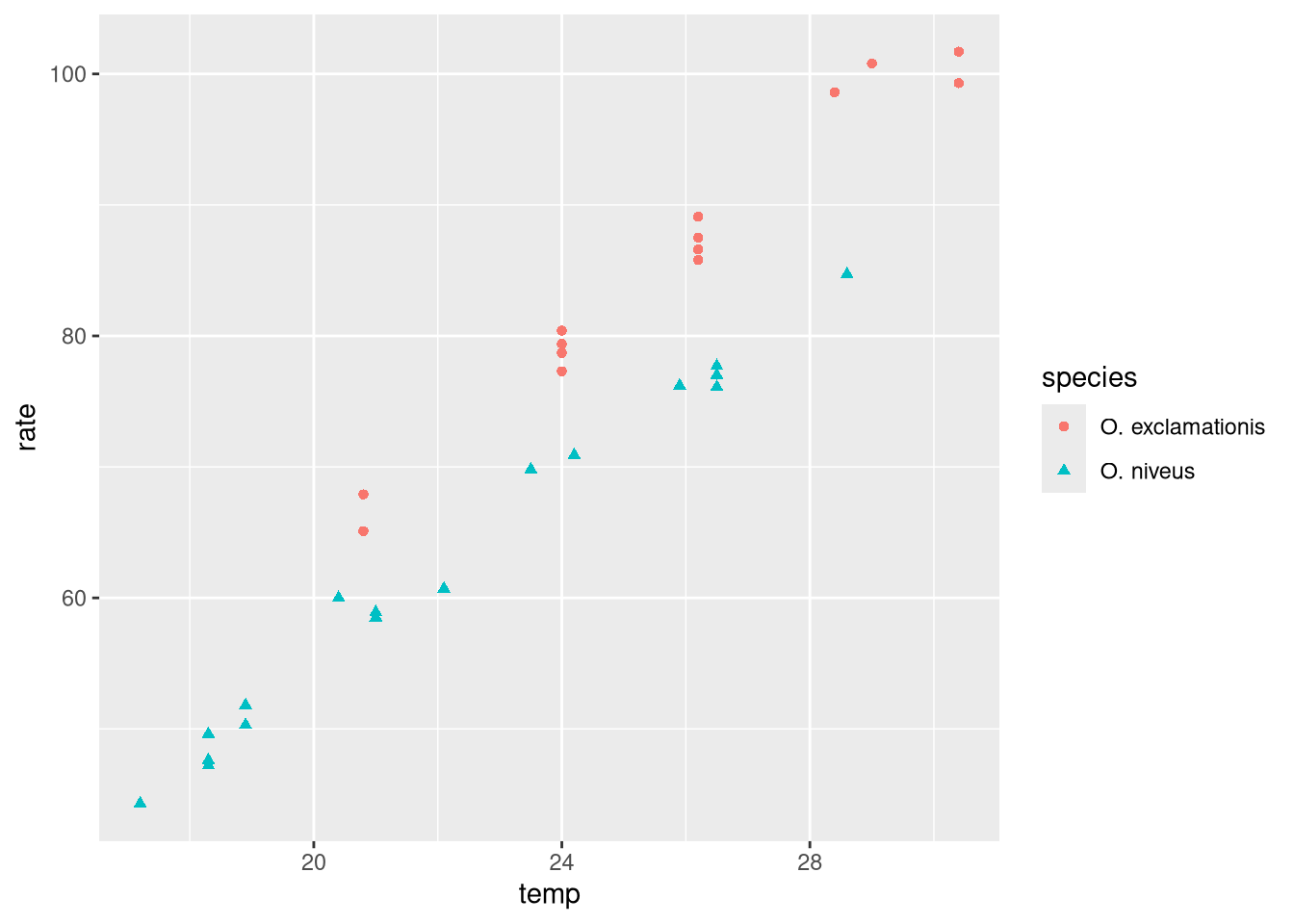
ggplot (data = crickets, aes (x = temp, y = rate, color = species,pch = species)) + geom_point ()
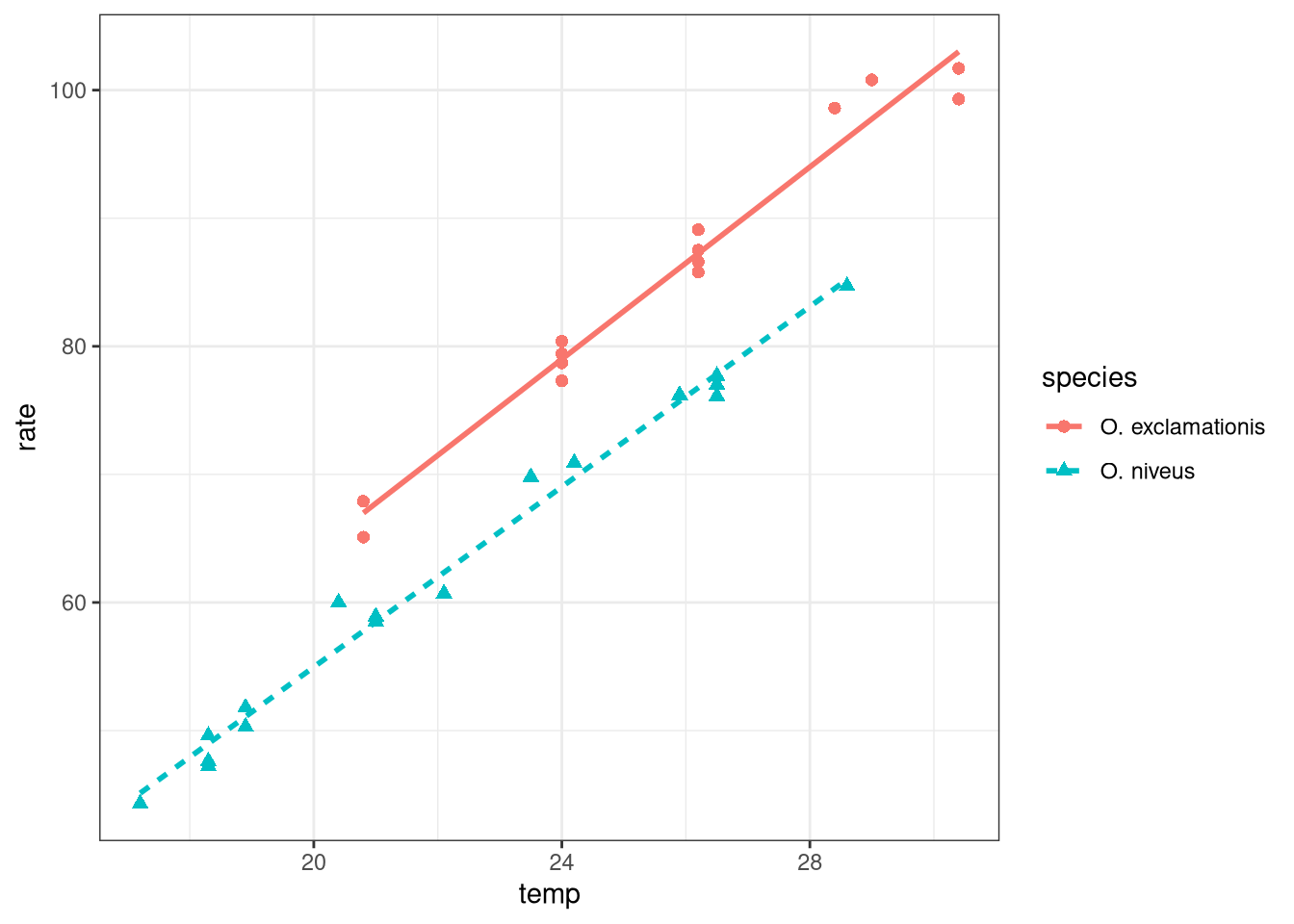
ggplot (data = crickets, aes (x = temp, y = rate, color = species,pch = species,lty = species)) + geom_point (size = 2 ) + geom_smooth (method = "lm" , se = FALSE , alpha = 0.5 ) + theme_bw ()
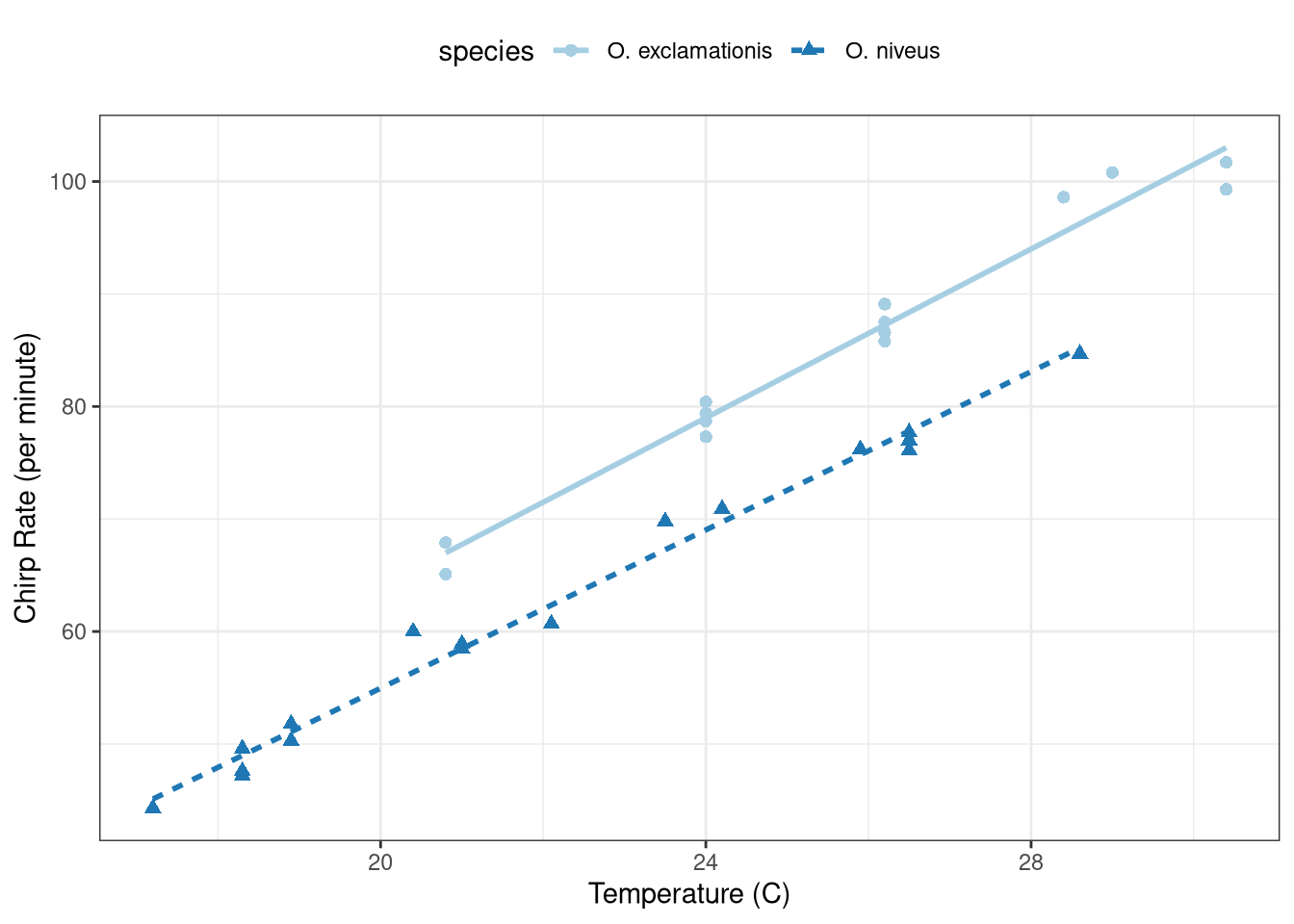
ggplot (data = crickets, aes (x = temp, y = rate, color = species,pch = species,lty = species)) + geom_point (size = 2 ) + geom_smooth (method = "lm" , se = FALSE , alpha = 0.5 ) + scale_color_brewer (palette = "Paired" ) + theme_bw () + labs (x = "Temperature (C)" , y = "Chirp Rate (per minute)" ) + theme (legend.position = "top" )
The variable species is stored in R as a factor. Most models need to encode the species data in a numeric format. The model formula rate ~ temp + species, will automatically encode the species column by adding a new numeric column that has a 0 when the species is O. exclamationis and a 1 when the species is O. niveus.
<- lm (rate ~ temp + species, data = crickets)model.matrix (mod_lm) |> bind_cols (SPECIES = crickets$ species)
# A tibble: 31 × 4
`(Intercept)` temp `speciesO. niveus` SPECIES
<dbl> <dbl> <dbl> <fct>
1 1 20.8 0 O. exclamationis
2 1 20.8 0 O. exclamationis
3 1 24 0 O. exclamationis
4 1 24 0 O. exclamationis
5 1 24 0 O. exclamationis
6 1 24 0 O. exclamationis
7 1 26.2 0 O. exclamationis
8 1 26.2 0 O. exclamationis
9 1 26.2 0 O. exclamationis
10 1 26.2 0 O. exclamationis
# ℹ 21 more rows
There are four levels to the variable heating. The model formula automatically adds three additional binary columns that are binary indicators for three of the heating types. The reference level of the factor (i.e., the first level) is omitted.
library (PASWR2) # For VIT2005 # heating has four levels <- lm (totalprice ~ area + heating, data = VIT2005)model.matrix (mod_example) |> bind_cols (HEATING = VIT2005$ heating)
# A tibble: 218 × 6
`(Intercept)` area heating3A heating3B heating4A HEATING
<dbl> <dbl> <dbl> <dbl> <dbl> <fct>
1 1 75.3 1 0 0 3A
2 1 101. 0 0 1 4A
3 1 88.9 1 0 0 3A
4 1 62.6 0 0 0 1A
5 1 146. 0 0 1 4A
6 1 77.2 1 0 0 3A
7 1 77.0 0 0 1 4A
8 1 62.9 1 0 0 3A
9 1 58.5 1 0 0 3A
10 1 59.2 1 0 0 3A
# ℹ 208 more rows
The model formula rate ~ temp + species creates a model with different y-intercepts for each species and the same slope.
Call:
lm(formula = rate ~ temp + species, data = crickets)
Coefficients:
(Intercept) temp speciesO. niveus
-7.211 3.603 -10.065
# The following may be easier to understand <- lm (rate ~ temp + species + 0 , data = crickets)
Call:
lm(formula = rate ~ temp + species + 0, data = crickets)
Coefficients:
temp speciesO. exclamationis speciesO. niveus
3.603 -7.211 -17.276
To get different slopes for each species, consider adding an interaction with one of the following:
<- lm (rate ~ temp + species + temp: species, data = crickets)
Call:
lm(formula = rate ~ temp + species + temp:species, data = crickets)
Coefficients:
(Intercept) temp speciesO. niveus
-11.041 3.751 -4.348
temp:speciesO. niveus
-0.234
<- lm (rate ~ temp* species, data = crickets)
Call:
lm(formula = rate ~ temp * species, data = crickets)
Coefficients:
(Intercept) temp speciesO. niveus
-11.041 3.751 -4.348
temp:speciesO. niveus
-0.234
<- lm (rate ~ (temp + species)^ 2 , data = crickets)
Call:
lm(formula = rate ~ (temp + species)^2, data = crickets)
Coefficients:
(Intercept) temp speciesO. niveus
-11.041 3.751 -4.348
temp:speciesO. niveus
-0.234
Write out the two separate lines from mod_int1.
\[\widehat{\text{rate}} = (-11.041) + (3.751)\cdot \text{temp}\]
\[\widehat{\text{rate}} = (-11.041 -4.348) + (3.751 - 0.234)\cdot \text{temp}\]
The output/models may be easier to understand with:
<- lm (rate ~ (temp + species + temp: species) + 0 , data = crickets)
Call:
lm(formula = rate ~ (temp + species + temp:species) + 0, data = crickets)
Coefficients:
temp speciesO. exclamationis speciesO. niveus
3.751 -11.041 -15.389
temp:speciesO. niveus
-0.234
Is the interaction term warranted?
<- lm (rate ~ temp + species,data = crickets)<- lm (rate ~ temp + species + temp: species,data = crickets)anova (mod_reduced, mod_full) |> tidy () -> ans|> :: kable ()
rate ~ temp + species
28
89.34987
NA
NA
NA
NA
rate ~ temp + species + temp:species
27
85.07409
1
4.275779
1.357006
0.2542464
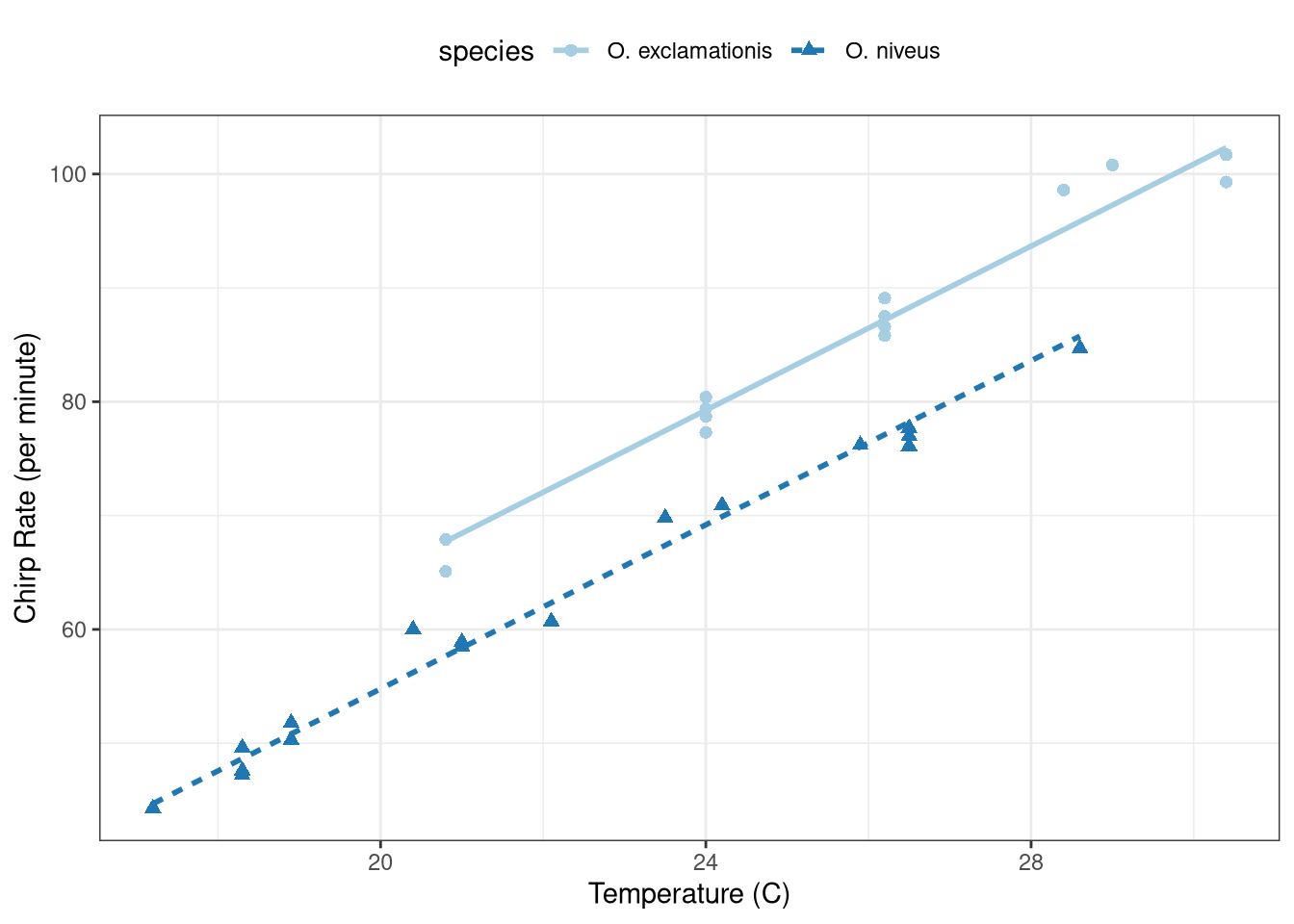
The p-value of 0.25 suggests \(\beta_3 = 0\) . The interaction term is not warranted and should be dropped from the model.
The model is a parallel slopes model and is shown in Figure 1 .
ggplot (data = crickets, aes (x = temp, y = rate, color = species,pch = species,lty = species)) + geom_point (size = 2 ) + :: geom_parallel_slopes (se = FALSE , alpha = 0.5 ) + scale_color_brewer (palette = "Paired" ) + theme_bw () + labs (x = "Temperature (C)" , y = "Chirp Rate (per minute)" ) + theme (legend.position = "top" )
Or using the coefficients from mod_lm:
Call:
lm(formula = rate ~ temp + species, data = crickets)
Coefficients:
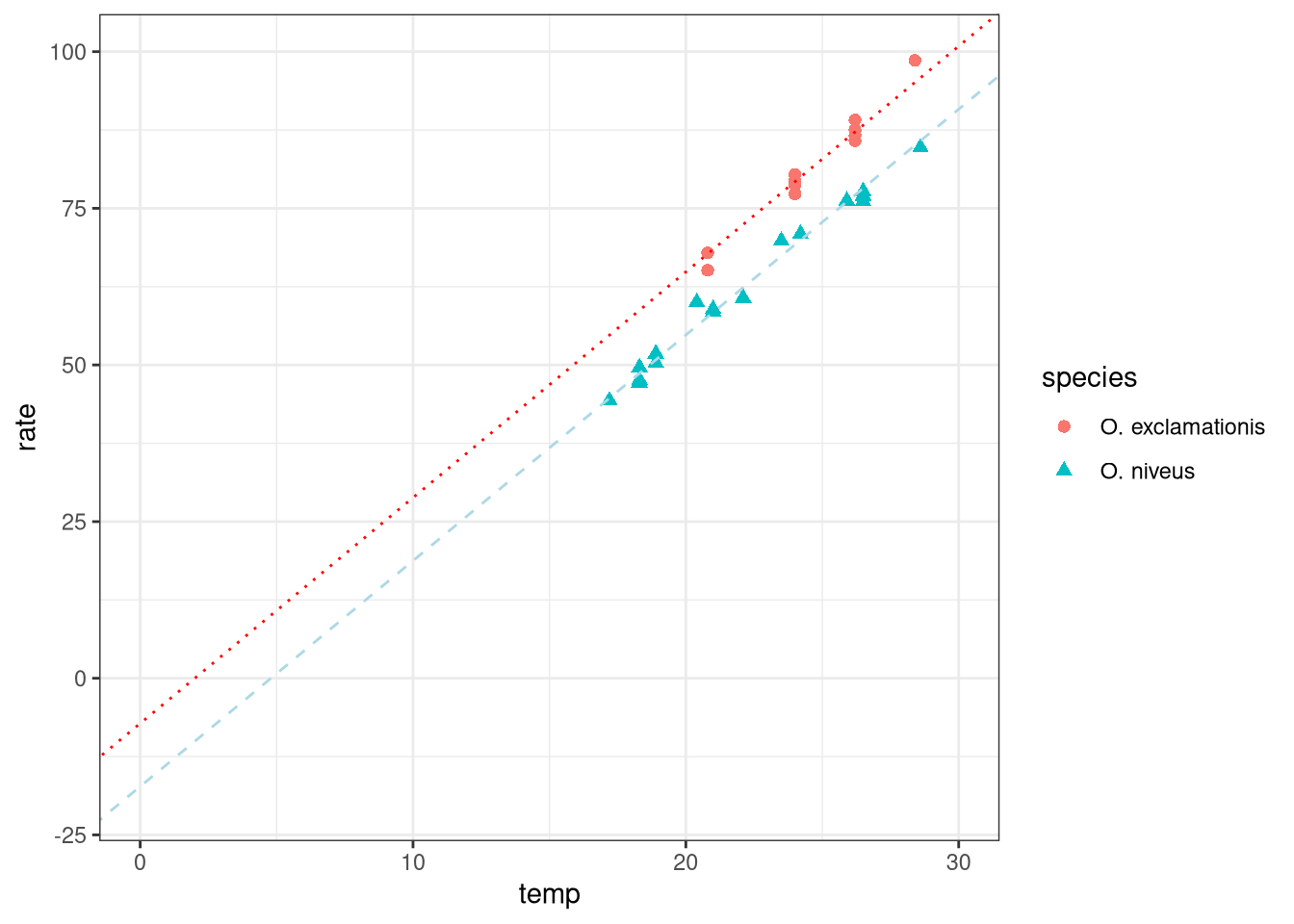
(Intercept) temp speciesO. niveus
-7.211 3.603 -10.065
ggplot (data = crickets, aes (x = temp, y = rate, color = species,pch = species,lty = species)) + geom_point (size = 2 ) + geom_abline (intercept = - 7.211 , slope = 3.603 , lty = "dotted" , color = "red" ) + geom_abline (intercept = - 17.276 , slope = 3.603 , lty = "dashed" , color = "lightblue" ) + theme_bw () + xlim (0 , 30 ) + ylim (- 20 , 100 )
cyl disp hp drat wt qsec vs am gear carb
Mazda RX4 6 160.0 110 3.90 2.620 16.46 0 1 4 4
Mazda RX4 Wag 6 160.0 110 3.90 2.875 17.02 0 1 4 4
Datsun 710 4 108.0 93 3.85 2.320 18.61 1 1 4 1
Hornet 4 Drive 6 258.0 110 3.08 3.215 19.44 1 0 3 1
Hornet Sportabout 8 360.0 175 3.15 3.440 17.02 0 0 3 2
Valiant 6 225.0 105 2.76 3.460 20.22 1 0 3 1
Duster 360 8 360.0 245 3.21 3.570 15.84 0 0 3 4
Merc 240D 4 146.7 62 3.69 3.190 20.00 1 0 4 2
Merc 230 4 140.8 95 3.92 3.150 22.90 1 0 4 2
Merc 280 6 167.6 123 3.92 3.440 18.30 1 0 4 4
Merc 280C 6 167.6 123 3.92 3.440 18.90 1 0 4 4
Merc 450SE 8 275.8 180 3.07 4.070 17.40 0 0 3 3
Merc 450SL 8 275.8 180 3.07 3.730 17.60 0 0 3 3
Merc 450SLC 8 275.8 180 3.07 3.780 18.00 0 0 3 3
Cadillac Fleetwood 8 472.0 205 2.93 5.250 17.98 0 0 3 4
Lincoln Continental 8 460.0 215 3.00 5.424 17.82 0 0 3 4
Chrysler Imperial 8 440.0 230 3.23 5.345 17.42 0 0 3 4
Fiat 128 4 78.7 66 4.08 2.200 19.47 1 1 4 1
Honda Civic 4 75.7 52 4.93 1.615 18.52 1 1 4 2
Toyota Corolla 4 71.1 65 4.22 1.835 19.90 1 1 4 1
Toyota Corona 4 120.1 97 3.70 2.465 20.01 1 0 3 1
Dodge Challenger 8 318.0 150 2.76 3.520 16.87 0 0 3 2
AMC Javelin 8 304.0 150 3.15 3.435 17.30 0 0 3 2
Camaro Z28 8 350.0 245 3.73 3.840 15.41 0 0 3 4
Pontiac Firebird 8 400.0 175 3.08 3.845 17.05 0 0 3 2
Fiat X1-9 4 79.0 66 4.08 1.935 18.90 1 1 4 1
Porsche 914-2 4 120.3 91 4.43 2.140 16.70 0 1 5 2
Lotus Europa 4 95.1 113 3.77 1.513 16.90 1 1 5 2
Ford Pantera L 8 351.0 264 4.22 3.170 14.50 0 1 5 4
Ferrari Dino 6 145.0 175 3.62 2.770 15.50 0 1 5 6
Maserati Bora 8 301.0 335 3.54 3.570 14.60 0 1 5 8
Volvo 142E 4 121.0 109 4.11 2.780 18.60 1 1 4 2
:: map (mtcars |> select (- mpg), cor.test, y = mtcars$ mpg)
$cyl
Pearson's product-moment correlation
data: .x[[i]] and mtcars$mpg
t = -8.9197, df = 30, p-value = 6.113e-10
alternative hypothesis: true correlation is not equal to 0
95 percent confidence interval:
-0.9257694 -0.7163171
sample estimates:
cor
-0.852162
$disp
Pearson's product-moment correlation
data: .x[[i]] and mtcars$mpg
t = -8.7472, df = 30, p-value = 9.38e-10
alternative hypothesis: true correlation is not equal to 0
95 percent confidence interval:
-0.9233594 -0.7081376
sample estimates:
cor
-0.8475514
$hp
Pearson's product-moment correlation
data: .x[[i]] and mtcars$mpg
t = -6.7424, df = 30, p-value = 1.788e-07
alternative hypothesis: true correlation is not equal to 0
95 percent confidence interval:
-0.8852686 -0.5860994
sample estimates:
cor
-0.7761684
$drat
Pearson's product-moment correlation
data: .x[[i]] and mtcars$mpg
t = 5.096, df = 30, p-value = 1.776e-05
alternative hypothesis: true correlation is not equal to 0
95 percent confidence interval:
0.4360484 0.8322010
sample estimates:
cor
0.6811719
$wt
Pearson's product-moment correlation
data: .x[[i]] and mtcars$mpg
t = -9.559, df = 30, p-value = 1.294e-10
alternative hypothesis: true correlation is not equal to 0
95 percent confidence interval:
-0.9338264 -0.7440872
sample estimates:
cor
-0.8676594
$qsec
Pearson's product-moment correlation
data: .x[[i]] and mtcars$mpg
t = 2.5252, df = 30, p-value = 0.01708
alternative hypothesis: true correlation is not equal to 0
95 percent confidence interval:
0.08195487 0.66961864
sample estimates:
cor
0.418684
$vs
Pearson's product-moment correlation
data: .x[[i]] and mtcars$mpg
t = 4.8644, df = 30, p-value = 3.416e-05
alternative hypothesis: true correlation is not equal to 0
95 percent confidence interval:
0.4103630 0.8223262
sample estimates:
cor
0.6640389
$am
Pearson's product-moment correlation
data: .x[[i]] and mtcars$mpg
t = 4.1061, df = 30, p-value = 0.000285
alternative hypothesis: true correlation is not equal to 0
95 percent confidence interval:
0.3175583 0.7844520
sample estimates:
cor
0.5998324
$gear
Pearson's product-moment correlation
data: .x[[i]] and mtcars$mpg
t = 2.9992, df = 30, p-value = 0.005401
alternative hypothesis: true correlation is not equal to 0
95 percent confidence interval:
0.1580618 0.7100628
sample estimates:
cor
0.4802848
$carb
Pearson's product-moment correlation
data: .x[[i]] and mtcars$mpg
t = -3.6157, df = 30, p-value = 0.001084
alternative hypothesis: true correlation is not equal to 0
95 percent confidence interval:
-0.7546480 -0.2503183
sample estimates:
cor
-0.5509251
<- purrr:: map (mtcars |> select (- mpg), cor.test, y = mtcars$ mpg)10 ]]
Pearson's product-moment correlation
data: .x[[i]] and mtcars$mpg
t = -3.6157, df = 30, p-value = 0.001084
alternative hypothesis: true correlation is not equal to 0
95 percent confidence interval:
-0.7546480 -0.2503183
sample estimates:
cor
-0.5509251
10 ]] |> :: tidy ()
# A tibble: 1 × 8
estimate statistic p.value parameter conf.low conf.high method alternative
<dbl> <dbl> <dbl> <int> <dbl> <dbl> <chr> <chr>
1 -0.551 -3.62 0.00108 30 -0.755 -0.250 Pearson's… two.sided
|> map_dfr (tidy, .id = "predictor" ) -> output |> :: kable ()
cyl
-0.8521620
-8.919699
0.0000000
30
-0.9257694
-0.7163171
Pearson’s product-moment correlation
two.sided
disp
-0.8475514
-8.747151
0.0000000
30
-0.9233594
-0.7081376
Pearson’s product-moment correlation
two.sided
hp
-0.7761684
-6.742388
0.0000002
30
-0.8852686
-0.5860994
Pearson’s product-moment correlation
two.sided
drat
0.6811719
5.096042
0.0000178
30
0.4360484
0.8322010
Pearson’s product-moment correlation
two.sided
wt
-0.8676594
-9.559044
0.0000000
30
-0.9338264
-0.7440872
Pearson’s product-moment correlation
two.sided
qsec
0.4186840
2.525213
0.0170820
30
0.0819549
0.6696186
Pearson’s product-moment correlation
two.sided
vs
0.6640389
4.864385
0.0000342
30
0.4103630
0.8223262
Pearson’s product-moment correlation
two.sided
am
0.5998324
4.106127
0.0002850
30
0.3175583
0.7844520
Pearson’s product-moment correlation
two.sided
gear
0.4802848
2.999191
0.0054009
30
0.1580618
0.7100628
Pearson’s product-moment correlation
two.sided
carb
-0.5509251
-3.615750
0.0010844
30
-0.7546480
-0.2503183
Pearson’s product-moment correlation
two.sided
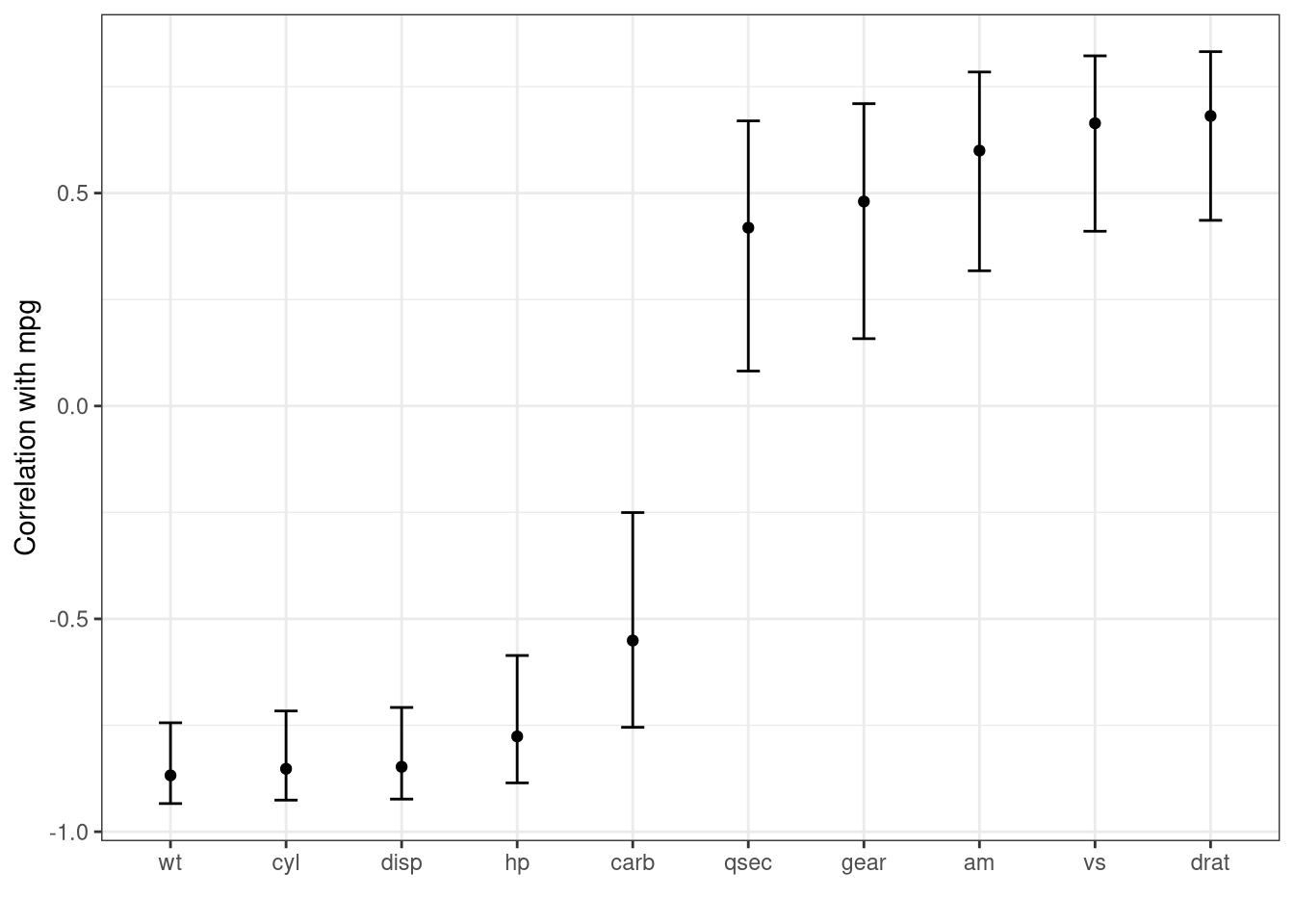
Create a graph with output.
ggplot (data = output, aes (x = fct_reorder (predictor, estimate))) + geom_point (aes (y = estimate)) + geom_errorbar (aes (ymin = conf.low, ymax = conf.high), width = 0.2 ) + labs (x = "" , y = "Correlation with mpg" ) + theme_bw ()
Combing Base R Models and Tidyverse
|> group_nest (species) |> :: kable ()
O. exclamationis
20.8, 20.8, 24.0, 24.0, 24.0, 24.0, 26.2, 26.2, 26.2, 26.2, 28.4, 29.0, 30.4, 30.4, 67.9, 65.1, 77.3, 78.7, 79.4, 80.4, 85.8, 86.6, 87.5, 89.1, 98.6, 100.8, 99.3, 101.7
O. niveus
17.2, 18.3, 18.3, 18.3, 18.9, 18.9, 20.4, 21.0, 21.0, 22.1, 23.5, 24.2, 25.9, 26.5, 26.5, 26.5, 28.6, 44.3, 47.2, 47.6, 49.6, 50.3, 51.8, 60.0, 58.5, 58.9, 60.7, 69.8, 70.9, 76.2, 76.1, 77.0, 77.7, 84.7
|> group_nest (species) |> mutate (model = map (data, ~ lm (rate ~ temp, data = .x)))
# A tibble: 2 × 3
species data model
<fct> <list<tibble[,2]>> <list>
1 O. exclamationis [14 × 2] <lm>
2 O. niveus [17 × 2] <lm>
# |> group_nest (species) |> mutate (model = map (data, ~ lm (rate ~ temp, data = .x))) |> mutate (coef = map (model, tidy))
# A tibble: 2 × 4
species data model coef
<fct> <list<tibble[,2]>> <list> <list>
1 O. exclamationis [14 × 2] <lm> <tibble [2 × 5]>
2 O. niveus [17 × 2] <lm> <tibble [2 × 5]>
# |> group_nest (species) |> mutate (model = map (data, ~ lm (rate ~ temp, data = .x))) |> mutate (coef = map (model, tidy)) |> select (species, coef) |> unnest (cols = c (coef))
# A tibble: 4 × 6
species term estimate std.error statistic p.value
<fct> <chr> <dbl> <dbl> <dbl> <dbl>
1 O. exclamationis (Intercept) -11.0 4.77 -2.32 3.90e- 2
2 O. exclamationis temp 3.75 0.184 20.4 1.10e-10
3 O. niveus (Intercept) -15.4 2.35 -6.56 9.07e- 6
4 O. niveus temp 3.52 0.105 33.6 1.57e-15